What type of tools would be needed to build copilot for pre-surgery screening? And how would they be integrated together?
Let’s review the problem we’re solving, and describe a few possible approaches and solutions. This is a project Analytiq Hub developed for Boston Medical Data. ![]()
The Pre-Surgery Screening Task
Below are the steps completed by a nurse for pre-surgery screening:
- The nurse needs to review patient allergies, medical conditions, tests, previous surgeries and medications.
- The patient is called by the nurse, who reviews the required information verbally.
- The nurse generates a report of the screening call.
The input comes in form of:
- Patient consent form
- Doctor progress notes, summarizing recent patient appointments
- Blood tests
- ECG, other labs
- Pre-op exam notes
Design components
The design includes three steps:
- Document Preparation
- Call Transcription
- Call Summarization
In a bit more detail:
- Document Preparation will scan the patient consent form, doctor progress notes, tests and labs, and prepare a call script, summarizing essential information that needs to be obtained during the call.
- Call Transcription will record the call, and transcribe it to text.
- Call Summarization will take as input the initial documents and the call transcript, and will generate a summary.
In the 1st part of our post series, we describe the Document Preparation part of the implementation.
Medical Screening Call Document Preparation
Preparation can be implemented with the following services:
- AWS Textract will OCR the input PDF. The output will be a JSON block consisting of all text elements and their relationships (e.g., which word is on which line, and which line is on which page). We extract these blocks further into per-page text, and into tables.
- AWS Comprehend Medical is used to anonymize the extracted text pages. Patient name, birth date, and any identifiable information is removed or replaced.
- OpenAI GPT4 is used on the anonymized text pages, to extract patient allergies, medical conditions, and medications.
- OpenAI GPT4 is again used to merge the per-page output and remove duplicates. This is used to generate the call input summary.
The anonymization step is not necessary if we enter a Business Associate Agreement with OpenAI, to ensure HIPAA compliance – or if, instead of GPT4, we use a language model that is part of AWS Bedrock.
The architecture diagram is:
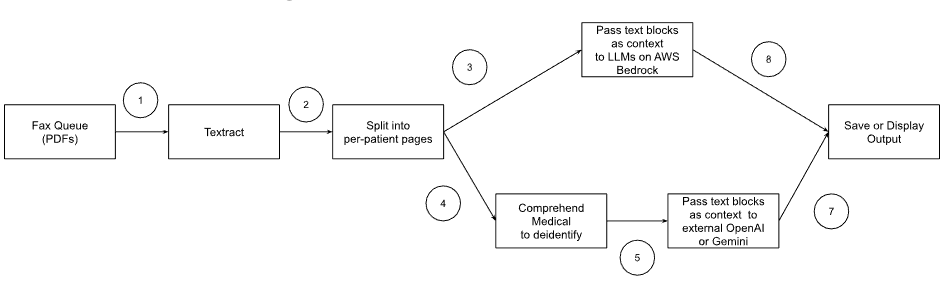
- OCR for the fax PDF
- Textract has accurate enough OCR for the three sample faxes provided
- Alternative parsers: Azure OCR, Tesseract, PaddleOCR
- Alternative: Donut for combined OCR+extraction
- Output of Textract shows which words are on which page. Shows tables and key/value pairs.
- Words are accurately detected for the most part.
- Tables sometimes have neighboring cells mistakenly ‘merged’ during parsing
- Key/value pairs are useful but not always reliable.
- Tables and key/value pairs could be used if the documents had a predictable format. In our case, the PDFs are pretty irregular.
- There is enough in the Textract output to be able to separate pages per patient.
- Use AWS Bedrock
- Supports Claude, LLaMa2, AWS Titan (fine tunable) but not OpenAI.
- LLM context window needs to fit the text from each PDF page.
- We prompt the LLM for allergies, medical conditions, medications for each page.
- Then, assemble the output for all the pages, prompting again to remove duplicates.
- AWS Bedrock allows fine tuning for AWS Titan. That could improve performance considerably.
- Given AWS Bedrock is on-prem, this can be done w/o deidentification
- Deidentification: Run AWS Medical Comprehend on each page.
- Goal is merely to find which text tokens need to be identify
- Generate deidentified records.
- AWS Medical Comprehend runs on text input. Textract generates fancy relationships in its output. All those relationships would be discarded, except for the page number.
- Same as 3, but using deidentified data – which can be passed to GPT4, Anthropic, Cohere or Gemini LLMs
- If BAA agreement with LLM vendor, no need to deidentify data – and can skip step 4
- Advantage of GPT4 is – better results
- OpenAI allows fine tuning of data for GPT3. That could improve performance considerably, b/c our output is easily measured for correctness.
In this blog post, we keep the anonymization step, and we use the GPT4 language model.
Here is a way to implement this. We import the required python modules:
import boto3
import os
import sys
import re
import json
import time
from collections import defaultdictThis routine runs AWS Textract in async mode. Our documents are too large and would fail in Textract if we use instead Textract sync mode:
def get_textract_blocks(s3_bucket_name, s3_key,
query_list=None):
# process using image bytes
session = boto3.Session()
textract = session.client('textract')
# Start the asynchronous document analysis
if query_list is not None and len(query_list) > 0:
query_list = [{'Text': '{}'.format(q)} for q in query_list]
response = textract.start_document_analysis(
DocumentLocation={
'S3Object': {
'Bucket': s3_bucket_name,
'Name': s3_key
}
},
FeatureTypes=["TABLES", "FORMS", "QUERIES"],
QueriesConfig = {'Queries': query_list}
)
else:
response = textract.start_document_analysis(
DocumentLocation={
'S3Object': {
'Bucket': s3_bucket_name,
'Name': s3_key
}
},
FeatureTypes=["TABLES", "FORMS"],
)
# Get the Job ID
job_id = response['JobId']
# Continuously check the job status
idx = 0
while True:
status_response = textract.get_document_analysis(JobId=job_id)
status = status_response['JobStatus']
print(f"Job status {idx}: {status}")
idx += 1
if status in ["SUCCEEDED", "FAILED"]:
break
time.sleep(5) # Wait for 5 seconds before checking the job status again
# If the job succeeded, process the results
if status == "SUCCEEDED":
blocks = []
# Iterating over the results
next_token = None
while True:
# Adding a next_token to the request if more results are available
if next_token:
response = textract.get_document_analysis(JobId=job_id, NextToken=next_token)
else:
response = textract.get_document_analysis(JobId=job_id)
blocks.extend(response['Blocks'])
# Check if there are more results
next_token = response.get('NextToken', None)
if not next_token:
break
return blocks
# Handle the case where the job failed
else:
raise ValueError("Document analysis failed")
In our implementation, we also extract tables, forms, and queries. However, forms and queries are not really that useful for our use case. When productizing this solution, it would be preferable to disable forms and queries – and, actually, disable tables as well. Tables are usually well parsed by Textract, however, in our case, tables can have different formats, and the unpredictability of the format makes it hard to parse Textract table output, even if the tables are parsed correctly.
This routine reorganizes the parsed Textract blocks as a hash indexed by ID. This makes it more efficient to look up blocks:
def get_block_map(blocks):
block_map = {}
for block in blocks:
block_id = block['Id']
block_map[block_id] = block
return block_mapWe define a few more utilities that parse Textract blocks:
def get_kv_map(blocks):
# get key and value maps
key_map = {}
value_map = {}
for block in blocks:
block_id = block['Id']
if block['BlockType'] == "KEY_VALUE_SET":
if 'KEY' in block['EntityTypes']:
key_map[block_id] = block
else:
value_map[block_id] = block
return key_map, value_map
def get_kv_relationship(key_map, value_map, block_map):
kvs = defaultdict(list)
for _, key_block in key_map.items():
value_block = find_value_block(key_block, value_map)
key = get_text(key_block, block_map)
val = get_text(value_block, block_map)
kvs[key].append(val)
return kvs
def find_value_block(key_block, value_map):
for relationship in key_block['Relationships']:
if relationship['Type'] == 'VALUE':
for value_id in relationship['Ids']:
value_block = value_map[value_id]
return value_blockThis routine converts Textract blocks into text. It is implemented in such a way as to capture checkboxes in input documents. Sometimes, medical documents use checkboxes to mark certain conditions or medical procedures:
def get_text(block, blocks_map):
text = ''
if 'Relationships' in block:
for relationship in block['Relationships']:
if relationship['Type'] == 'CHILD':
for child_id in relationship['Ids']:
word = blocks_map[child_id]
if word['BlockType'] == 'WORD':
text += word['Text'] + ' '
if word['BlockType'] == 'SELECTION_ELEMENT':
if word['SelectionStatus'] == 'SELECTED':
text += 'X '
return textMore utilities:
def get_query_map(block_map):
query_map = {}
# Iterate through the block_map
for _, block in block_map.items():
if block["BlockType"] == "QUERY":
query_text = block["Query"]["Text"]
query_map[query_text] = None
for relationship in block.get("Relationships", []):
if relationship["Type"] == "ANSWER":
for value_id in relationship["Ids"]:
value_block = block_map[value_id]
answer_text = value_block["Text"]
query_map[query_text] = answer_text
return query_map
def print_kvs(kvs):
for key, value in kvs.items():
print(key, ":", value)
def search_value(kvs, search_key):
for key, value in kvs.items():
if re.search(search_key, key, re.IGNORECASE):
return valueSet up the s3 bucket and path, and set up the output:
s3_bucket_name = 'BUCKET_NAME' # Replace with the name of the s3 bucket storing the input PDF
s3_key = 'FILENAME.pdf' # The s3 path to the PDF.
# The PDF, in our case, is a concatenation of all the input documents.
# If the documents don't come into a single PDF, then the code below needs to be correspondingly changed.
output_dir = "OUTPUT_DIR" # Replace with a local disk directory
basename = os.path.basename(s3_key)
stem = os.path.splitext(basename)[0]Run textract:
blocks = get_textract_blocks(s3_bucket_name, s3_key,
query_list = [
"Patient birth date",
"Patient Name",
"Patient DOB",
"DOB",
"Patient age",
"allergies",
"Conditions",
"Proposed procedure",
"Medical record number",
"MRN"
])
block_map = get_block_map(blocks)This is the expected output:
Job status 0: IN_PROGRESS
Job status 1: IN_PROGRESS
Job status 2: IN_PROGRESS
Job status 3: SUCCEEDEDSave the blocks:
fname = f"{output_dir}/{stem}.json"
with open(fname, 'w') as f:
json.dump(blocks, f, indent=2)Save the text:
fname = f"{output_dir}/{stem}.txt"
with open(fname, 'w') as f:
for block in blocks:
if block['BlockType'] == 'LINE':
f.write(block['Text'] + "\n")Get the query map:
query_map = get_query_map(block_map)Get the key value map:
key_map, value_map = get_kv_map(blocks)
kvs = get_kv_relationship(key_map, value_map, block_map)Save the key value map:
fname = f"{output_dir}/{stem}_kvs.json"
with open(fname, "w") as f:
json.dump(kvs, f, indent=2)Utility to get the tables:
def get_tables(block_map):
tables = []
for _, block in block_map.items():
if block['BlockType'] == 'TABLE':
table = {}
for relationship in block.get('Relationships', []):
if relationship['Type'] == 'CHILD':
for child_id in relationship['Ids']:
cell = next(b for _, b in block_map.items() if b['Id'] == child_id)
row_index = cell['RowIndex']
col_index = cell['ColumnIndex']
text = ''
# Extracting the text from the cell
for relationship2 in cell.get('Relationships', []):
if relationship2['Type'] == 'CHILD':
for child_id2 in relationship2['Ids']:
child_block2 = block_map.get(child_id2, None)
if child_block2 and 'Text' in child_block2:
if text == "":
text = child_block2['Text']
else:
text += " "
text += child_block2['Text']
# Comment in to print the cell as json
# print(json.dumps(cell))
# Comment in to print the text in the cell
#print(text)
# Save the row and column index to the cell
table.setdefault(row_index, {})[col_index] = text
# Save the table to the list of tables
tables += [table]
# Comment in to print the table
#for row in sorted(table.keys()):
# print([table[row].get(col, '') for col in sorted(table[row].keys())])
return tablesGet the tables:
tables = get_tables(block_map)
# Save the tables
fname = f"{output_dir}/{stem}_tables.txt"
with open(fname, "w") as f:
for table in tables:
for row in sorted(table.keys()):
f.write(str([table[row].get(col, '') for col in sorted(table[row].keys())]))
f.write("\n")
f.write("\n")Construct the page text map:
def get_page_text_map(block_map):
page_text_map = {}
for _, block in block_map.items():
# We could parse the PAGE block types, read the LINE relationships
# Or we could parse the TEXT block types, and read the PAGE relationships.
# We do the latter.
if block['BlockType'] == 'LINE':
page = block['Page']
if page not in page_text_map:
page_text_map[page] = ""
page_text_map[page] += block['Text'] + " "
return page_text_map
page_text_map = get_page_text_map(block_map)Get the Comprehend Medical handle:
comprehend_medical_client = boto3.client('comprehendmedical')
Run Comprehend Medical and save the output:
response_map = {}
for page_idx, page_text in page_text_map.items():
# Use Comprehend Medical to analyze the extracted text
response_map[page_idx] = comprehend_medical_client.detect_entities_v2(
Text=page_text
)
for page_idx, response in response_map.items():
fname = f"{output_dir}/{stem}_comprehend_page_{page_idx}.json"
with open(fname, "w") as f:
f.write(json.dumps(response, indent=2))Get the anonymized output:
def get_page_text_map_anonymized(page_text_map, response_map):
page_text_map_anonymized = {}
for page_idx, page_text in page_text_map.items():
page_text_map_anonymized[page_idx] = page_text
for entity in response_map[page_idx]['Entities']:
if entity['Category'] == 'PROTECTED_HEALTH_INFORMATION':
start = entity['BeginOffset']
end = entity['EndOffset']
# Create a string of the same length as the entity
# Replace all characters with "X" except for newlines
# This is to preserve the line breaks in the text
replace_text = "X" * (end - start)
for i in range(start, end):
if page_text_map_anonymized[page_idx][i] == '\n':
replace_text = replace_text[:i-start] + '\n' + replace_text[i-start+1:]
page_text_map_anonymized[page_idx] = page_text_map_anonymized[page_idx][:start] + replace_text + page_text_map_anonymized[page_idx][end:]
return page_text_map_anonymized
page_text_map_anonymized = get_page_text_map_anonymized(page_text_map, response_map)Set up and run an OpenAI prompt to extract allergies, medical conditions and medications from the anonymized output:
from openai import OpenAI
openai_client = OpenAI()
def query_openai(system_prompt, user_prompt, model="gpt-4"):
response = openai_client.chat.completions.create(
model=model,
messages=[
{"role": "system", "content": system_prompt},
{"role": "user", "content": user_prompt},
]
)
# Check that the respomse is valid
if response.object != "chat.completion":
raise ValueError(f"Invalid response from OpenAI: {response}")
# Check that the response is JSON
response_content = response.choices[0].message.content
try:
response_json = json.loads(response_content)
except:
print(f"Page {page_idx} invalid JSON from OpenAI: {response_content}")
response_json = {}
#raise ValueError(f"Invalid JSON from OpenAI: {response_content}")
return response_json
system_prompt = \
"""You are a doctor reviewing patient notes ahead of surgery. Respond briefly and completely to the questios."""
user_prompt_prefix = \
"""
In the anonymized medical notes below, what are the allergies, past medical history, and medications?
Report only patient medical conditions. Do not report conditions of other people.
Use this JSON format. Only output JSON. Do not include any other text in your response. Output empty json in case of no answer.
{
"Allergies": ["allergy1", "allergy2"],
"Medical conditions": ["condition1", "condition2"],
"Medications": ["medication1", "medication2"]
}
"""
def get_amm_json_map(system_prompt, user_prompt_prefix, page_text_map_anonymized):
amm_map = {}
# Use OpenAI to generate a response. Use gpt4.
for page_idx, page_text in page_text_map_anonymized.items():
amm_map[page_idx] = query_openai(system_prompt, user_prompt_prefix + "\n\n" + page_text)
return amm_map
amm_map = get_amm_json_map(system_prompt, user_prompt_prefix, page_text_map_anonymized)This gives per-page responses. Merge these into a single response:
def get_merged_response(response_json_map):
# Merge the responses
response_json = {}
for page_idx, response in response_json_map.items():
for key in response:
if key not in response_json:
response_json[key] = []
response_json[key] += response[key]
return response_json
amm_json = get_merged_response(amm_map)Deduplicate the merged responses:
def deduplicate_amm_json(amm_json):
user_prompt = \
"""
In the anonymized medical notes below, what are the allergies, past medical history, and medications?
Deduplicate any entries. For example, if the same allergy is listed twice, even with slightly different names, only list it once.
Keep only names that make medical sense. For example, if the name is "Tylenol", keep it. If the name is "pain medication", remove it.
Use this JSON format. Only output JSON. Do not include any other text in your response. Output empty json in case of no answer.
{
"Allergies": ["allergy1", "allergy2"],
"Medical conditions": ["condition1", "condition2"],
"Medications": ["medication1", "medication2"]
}
===
"""
user_prompt += "Medical Notes:\n"
user_prompt += f" Allergies: {amm_json.get('Allergies', [])}\n"
user_prompt += f" Medical conditions: {amm_json.get('Medical conditions', [])}\n"
user_prompt += f" Medications: {amm_json.get('Medications', [])}\n"
#print(user_prompt)
amm_json = query_openai(system_prompt, user_prompt)
return amm_json
amm_json2 = deduplicate_amm_json(amm_json)Finally, save the deduplicated allergies, medical conditions, and medications as output. This is the final step.
# Save the AMM JSON
fname = f"{output_dir}/{stem}_amm.json"
with open(fname, "w") as f:
json.dump(amm_json2, f, indent=2)
fname = f"{output_dir}/{stem}_amm_nondeduplicated.json"
with open(fname, "w") as f:
json.dump(amm_json, f, indent=2)
